Are Eurocentric Genetic Databases Hampering Health Care?
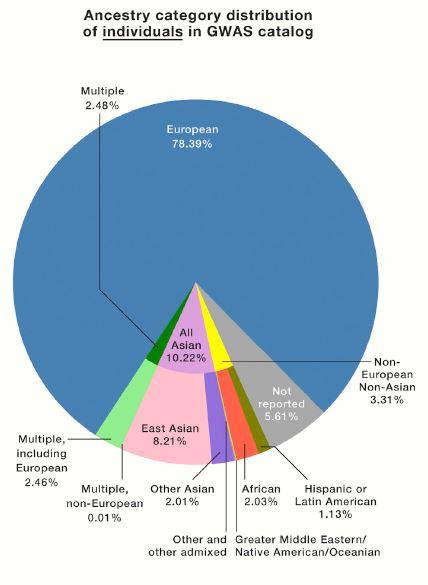
A commentary published today in the journal Cell offers evidence of a stunning imbalance in the population groups that participate in genetic and genomic research, which can underlie some health care inequities. A glance at the pie chart below, which represents many genome-wide association studies (GWAS), reveals a telling tipping in favor of participants of European ancestry – 78 percent. Asians account for just 10 percent, and Africans 2 percent.
A GWAS is a sweeping look at single sites (SNPs) among a genome’s 3.2 billion bases that differ among people. It’s used to trace traits and conditions caused by multiple genes plus environmental influences. Algorithms deduce the associations from SNP data from people who share ancestry as well as a trait or disease of interest. “Ancestry” means shared heritage, manifest as hunks of genomes, and not what a sociologist might call race based on appearance.
The Eurocentric nature of the pretty pie charts and polygons of consumer ancestry tests of course reflect the market. The skewed representation in company databases can lead to surprise, confusion, and disappointment, when a population simply isn’t yet on the radar.
In clinical studies, the stakes are higher. The overrepresentation of European whites can lead to sub-optimal diagnoses and therapeutic choices for people in other population groups. That’s what prompted the commentary in Cell, “The Missing Diversity in Human Genetic Studies,” from Giorgio Sirugo and Sarah Tishkoff from the University of Pennsylvania and Scott Williams of Case Western Reserve University. Citing examples from the long-studied single-gene conditions and others, they compellingly convey the importance of considering genetic ancestry in health care.
Eight Examples
-

(Carly Lewis) G6PD deficiency This may be the most common single-gene disease that most people in the U.S. haven’t heard of. It affects 400 million people worldwide and 200,000+ in the U.S., including 1 in 10 African American males (it’s transmitted on the X chromosome). The most serious symptom, hemolytic anemia, happens only under specific conditions: certain infections, use of certain drugs (anti-malarials, sulfonamides, aspirin, NSAIDs), or eating fava beans or inhaling fava pollen. It’s most prevalent among people who trace their ancestry to certain parts of Africa, Asia, the Mediterranean, and the Middle East. I had a genetic counseling patient who had severe anemia that several physicians had misdiagnosed because they hadn’t noticed her Greek surname and ordered the appropriate test. In Astoria, Queens, with its large Greek population, cans of fava beans on grocery store shelves bear warnings for people with the condition.
- BRCA cancers. “But 23andMe says I don’t have a mutation!” lamented a woman on a breast cancer Facebook page who indeed didn’t have the three most common mutations that the company checks. But when her provider sent a blood sample to clinical testing company Myriad Genetics, which considers many mutations and can sequence the entire gene, the findings changed. The consumer companies test only for the three “founder” mutations that are most prevalent among Ashkenazi Jewish people. But like a page in a book, a gene can have many different typos or missing words. Different populations have different frequencies of mutations in the same gene. Not everyone reads the abundant educational material on the consumer testing websites.
- Heart failure. One in ten cases of heart failure among African Americans is due to a specific dominant mutation in the ATTR-CM gene that’s found almost exclusively in this group; 4 percent are carriers. Because new treatments target the transthyretin protein that the gene encodes, diagnosing this form of heart disease right off the bat can save lives. In this form of heart failure (transthyretin amyloid cardiomyopathy), amyloid fibrils accumulate in the heart.
- Cystic fibrosis. Among white Europeans, 70% of cases are due to a double dose of the mutation F508del. But only 29% of African Americans with CF have it, and 12-31% of Asians with CF. No ancestral groups from Korea, Japan, Thailand,
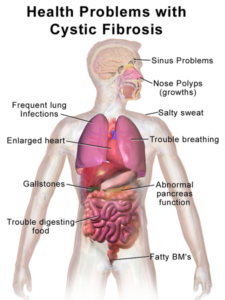
CF causes different sets of symptoms in different ancestral groups. Vietnam, or South Africa have the mutation. Because new CF treatments are targeted to mutations, identifying them, even if they’re rare, is important. It’s no surprise that CF is underdiagnosed among non-Europeans.
- LDL. The gene PCSK9 regulates receptors for low-density lipoproteins. A new class of “cholesterol busters” inhibits the PCSK9 protein and was developed with Europeans in mind. The common mutation among white Europeans with coronary heart disease due to high LDL from this genetic glitch leads to fewer receptors. LDL can’t get inside cells and spills into the bloodstream. Their mutation (called a “gain-of-function”) renders the gene overactive and the new drugs put on the brakes. But a different, “loss-of-function” mutation in the same gene among people of African ancestry has the opposite effect, leading to more receptors, lower blood LDL, and lower risk of coronary heart disease. A person of African ancestry with high LDL can likely blame a different gene than can a European. It wouldn’t make sense to prescribe a PCSK9 inhibitor.
6. Warfarin prescribing. This most commonly prescribed anti-coagulant works within a very narrow therapeutic window. Monitoring response to warfarin is important to balance clotting with bleeding. Algorithms based on variants of four genes that regulate dosage effects are the basis of monitoring tests. But those gene variants are predictive only for the Europeans on whom most of the studies have been based.
- Asthma. People of African, Mexican, and Puerto Rican ancestry have the most asthma and the poorest response to albuterol bronchodilators. A recent study sequenced the genomes of 1,440 children about equally split among Puerto Rican, Mexican, or African ancestry who either responded very well or very poorly to an inhaler. Leaving no stone unturned by considering entire genomes, the researchers discovered a gene controlling inhaler response in these kids. But they couldn’t replicate their findings because they couldn’t find other studies that had enough participants from these three groups. They conclude “The lack of minority data, despite a collaboration of eight universities and 13 individual laboratories, highlights the urgent need for a dedicated national effort to prioritize diversity in research.”
- Kidney failure. Two variants of the apolipoprotein L1 gene explain chronic and end stage kidney disease not associated with diabetes among African Americans, who face a 7 to 10-fold risk compared to people who don’t have either variant. Why is this form of kidney disease common among people of African ancestry but not others? Something that we geneticists call balancing selection. The variants of the encoded protein in people with mutations destroy the parasites that cause African sleeping sickness. So the mutations stick around, in carriers who survive sleeping sickness. It’s a variation on the theme of protection against malaria in carriers of sickle cell disease.
The History of the Problem
The Eurocentricity of DNA databases crept up on researchers, but it emerges in hindsight from the recent history of genomics.
In 1991, when the human genome project was beginning to make headlines, a different endeavor, headed by Stanford University geneticist Luigi Luca Cavalli-Sforza, sought to catalog the ways that genomes vary among people from ancestral populations all over the planet. These are the populations that founded a specific geographical area and stayed there, having children amongst themselves, so that chromosome hunks were passed down over generations with little if any change.
The resulting Human Genome Diversity Project (HGDP) was a response to the European-centric gene maps that were being used as a framework in the first sequencing efforts. “The focus of the HGDP was characterizing variation across populations. There was a backlash at the time over informed consent,” particularly among small, isolated populations being approached by researchers, Sarah Tishkoff told me.
Then the International HapMap Project began in 2002, just as the first human genome sequences were published. The goal was to consider genomes as blocks of SNPs, called haplotypes, that indicate ancestry (see last week’s post).
Next came tracking SNP blocks in genome-wide association studies (GWAS; biologists are terrible at acronyms). The first ones, reported in 2002, zeroed in on genome regions near the haplotypes that people with a distinctive trait or medical condition share. For years “GWAS” have peppered the tables of contents of genetics journals, persisting as short cuts while genome sequencing became more proficient. Today the two techniques work in tandem to find and focus on parts of genomes. The authors of the new paper consulted data from the GWAS Catalog through January 2019 to construct the pie charts that reveal the lack of diversity – confirming common knowledge.
GWAS used to find disease genes required large groups of patients, and that’s where the bias began, Tishkoff said. Perhaps sensitive to informed consent issues the HGDP raised, or concerned that statistics on a small group of people would be lost among those from much larger populations, reviewers of grant applications would sometimes request that a researcher omit a certain group from the proposed project, she added. It was just easier. But the omissions set the stage for today’s quiet crisis in diversity in DNA databases.
Another reality minimizing minority representation is that homogeneous populations, with little variation in the versions of different genes, are easier to study. Anomalies pop out. The boring sameness of European genomes is opposite the situation for African genomes, because Africans were the only humans for the first 220,000-or-so years of our 300,000-or-so years of existence.
And so the haplotypes of African genomes have splintered through time, with opportunity at each generation for chromosomes to exchange parts (cross over) as eggs and sperm formed and sometimes shuffled the pattern from the past. Once people began leaving Africa, the spectacular genome diversity became strangled through bottlenecks imposed by environmental and geographic challenges and by natural selection that favored some traits and jettisoned others. The genomes of Europeans and others are but subsets of ancestral African genomes, plus a few new mutations.
Diversifying DNA Databases
Fortunately, efforts are underway to make genetic research more inclusive. Researchers, from single lab groups to those at huge biobanks, are seeking participants from diverse population groups. Consumer genetic testing companies are bolstering reference populations, slowly diminishing the proportion of unsatisfied customers receiving false negative findings, and providing spit kits gratis to pharmaceutical companies who include underrepresented populations in drug discovery.
It’s time to divide up the pie of humanity to represent us all.
(I wrote a more technical version of this post for Medscape.)